
About CGB
Major achievements in basic research:
A large number of core-set microsatellite DNA markers have been developed and applied for cotton genome research. For the first time several DNA markers associated with cotton natural leaf defoliation were identified and localized on Ch18. Additionally genetic loci associated with fiber yield were identified and localized using chromosome substituted lines on chromosomes 12, 18, 23 and 26. Some DNA markers were also associated with fiber initiation and development and finally recommended for utilization in marker assisted breeding. DNA markers were successfully used for genetic mapping of photoperiod genes in cotton.
A large number of unique cotton small RNAs involved in initiation of cotton fiber development were cloned and annotated; small RNAs playing an important role in Fusarium wilt and root-knot nematode pathogenesis were also characterized.
For the first time in the world cotton genome research, the length of recombination blocks (i.e. the extent of linkage disequilibrium level) of cotton genome were estimated and DNA markers associated with fiber yield and quality were identified using association mapping. Molecular and phylogenetic diversity of cotton germplasm collection were studied.
A complex study of genetic structure of Uzbek population was done by using analysis of Alu insertions and mitochondrial HVR region polymorphisms.
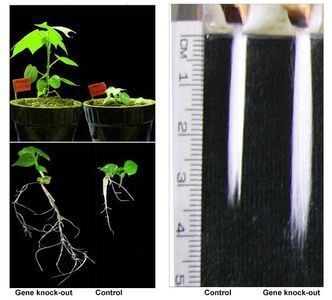
For the first time cotton phytochrome gene family were cloned and characterized, a molecular evolution of phytochrome gene family was studied. Phytochrome genes play an important role in photomorphogenesis, flowering and fiber elongation. It was shown that tetraploid cottons have four PHYA, two PHYB and two PHYE genes. We have developed phytochrome specific gene knock-out technology.
A number of DNA markers associated with cotton Fusarium wilt resistance were identified.
A family of cotton MIC-3 genes were cloned and sequenced. MIC-3 gene family plays an important role in plant defense along with PR-proteins. Molecular evolution of different cotton genomes was studied and for the first time pathogen-dependent gene duplication pattern in cotton was identified that completely fit with «bait and switch» and «guard and decoy» evolution models of resistant genes in plants.
From several diploid and tetraploid cotton genomes an ortholog of Arabidopsis eskimo-1 gene was cloned and sequenced, which is cold resistance gene and plays important role in drought resistance in cotton.
There is a research team of cotton genomics in CGB, which is successfully developing and keep on growing (academician Abdukarimov A. and Prof. Abdurakhmonov I.Y).
The scientists of the Center have received numerous international awards such as The World Academy of Sciences (TWAS, http://twas.ictp.it/) TWAS prize 2010, and ICAC researcher of the Year 2013 for fundamental contribution in the fields of cotton genomics, cotton genetic diversity, understanding of molecular basis of utilization of cotton diversity.
In 2012 a monography book on «Plant breeding» (ISBN 978-953-307-932-5, http://www.intechopen.com/books/plant-breeding) was published, which became a handbook for students and young researchers working on crop plants breeding.
In 2013, a special issue of «Cotton Research in Uzbekistan» in the Asian and Australasian Journal of Plant Sciences and Biotechnology (http://www.globalsciencebooks.info/Journals/AAJPSB.html) was published, where Uzbek scientists reported about current research and findings in the field of cotton science.
Applied research:
On the basis of fundamental studies of gene mapping , for the first time, a panel of core-set DNA markers effective for identification of loci associated with fiber quality and useful for marker assisted selection (MAS) programs was created. Using DNA-marker based technology, the fourth generations of MAS cotton lines with improved fiber quality were developed. These lines are being evaluated in field trials with the aim of commercialization of the first generation MAS cotton cultivars in Uzbekistan.
A molecular-genetic passports ( DNA barcodes) of cotton varieties and lines from Uzbek cotton germplasm collection were created which will be a basis for protection of intellectual property rights of cotton germplasm resources and genetically modified varieties developed in Uzbekistan.

For the first time, a gene knock-out technology in cotton was developed with successful silencing of genes responsible for flowering and fiber quality traits. Using this technology, a number of varieties (Porloq-1, 2, 3 and 4) with improved fiber quality, early maturity and high fiber yield have been developed.
In 2012 with the purpose of protecting the gene knock-out technology a patent applications for the concept of the technology have been filed in Uzbekistan (IAP20120069), USA (USPTO:13/445696) and internationally (PCT/US13/27801). For seed propagation of genetically modified cotton varieties, a special seed-breeding and production facility were built under support and supervision of Cabinet of Ministers of Uzbekistan.
With the aim of extension and further development of genomics science in Uzbekistan in CGB International Training Center was established, where students, young scientists and specialists are being routinely trained. There are a number of lecture series on novel genomic technologies have been prepared and published, which are being successfully used for educational process.
In CGB, GMO Testing Unit was established that has been accredited by National Agency of Standardization of Uzbekistan with the certificate № UZ.AMT.07.MAI. 736.